An Introduction to BayesHMM
Luis Damiano, Michael Weylandt, Brian Peterson
2019-01-03
Source:vignettes/introduction.Rmd
introduction.RmdBayesHMM is an R Package to run full Bayesian inference on Hidden Markov Models (HMM) using the probabilistic programming language Stan. By providing an intuitive, expressive yet flexible input interface, we enable researchers to profit the most out of the Bayesian workflow. We provide the user with an expressive interface to mix and match a wide array of options for the observation and latent models, including ample choices of densities, priors, and link functions whenever covariates are present. The software enables users to fit HMM with time-homogeneous transitions as well as time-varying transition probabilities. Priors can be set for every model parameter. Implemented inference algorithms include forward (filtering), forward-backwards (smoothing), Viterbi (most likely hidden path), prior predictive sampling, and posterior predictive sampling. Graphs, tables and other convenience methods for convergence diagnosis, goodness of fit, and data analysis are provided.
This vignette introduces the software and briefly reviews current capabilities. Although we walk the reader through most of the functionalities, we do not discuss function arguments, implementation details, and other details typically reviewed in the documentations. A list of future features may be found in the Roadmap wiki page.
NOTE: Our software is work in progress. We will review naming conventions as well as other design decisions at a later stage. Fully detailed documentation will be available at the time of release.
Using BayesHMM
The typical data analysis workflow includes the following steps: (1) specify, (2) validate, (3) simulate, (4) fit, (5) diagnose, (6) visualize, (7) compare. All steps except for 5 and 7 are implemented as of today.
You may install the lastest version of our software from GitHub. Please note that BayesHMM requires rstan (>= 2.17), the R interface for the probabilistic programming language Stan.
#> Loading required package: Rcpp
#> Hola! BayesHHM v0.0.1 here o/devtools::install_github("luisdamiano/BayesHMM", ref = "master")
library(BayesHMM)1. Specify
A HMM may be specified by calling the hmm function:
hmm(
K = 3, R = 2,
observation = {...}
initial = {...}
transition = {...}
name = "Model name"
)where \(K\) is the number of hidden states and \(R\) is the number of dimensions in the observation variable.
The observation, initial, and transition fields rely on S3 objects called Density to specify density form, parameter priors, and fixed values for parameters. User may specify bounds in the parameter space as well as truncation in prior densities. For example:
-
Gaussian(mu = 0, sigma = 1)specifies a Gaussian density with fixed parameters. -
Gaussian(mu = Gaussian(0, 10), sigma = Cauchy(0, 10, bounds = list(0, NA)))specifies a Gaussian density with a Gaussian prior on the location parameter and a Cauchy prior on the scale parameter, which is bounded on \([0, \infty)\).
Currently available densities are listed below:
- Observation model:
- Univariate: Bernoulli, Beta, Binomial, Categorical, Cauchy, Dirichlet, Gaussian, Multinomial, Negative Binomial (traditional and location parameterizations), Poisson, Student.
- Multivariate: Multivariate Gaussian (traditional, Cholesky decomposition of covariance matrix, and Cholesky decomposition of correlation matrix parameterizations), Multivariate Student.
- Regressions: Bernoulli regression with logit link, Binomial regression (logit and probit links), Softmax regression, Gaussian regression.
- Prior-only density: LKJ, Wishart.
- Transition model: Dirichlet, Softmax regression.
- Initial model: Dirichlet, Softmax regression.
Observation model
Internally, a specification stores either \(K\) multivariate densities (i.e. one multivariate density for state) or \(K \times R\) univariate densities (i.e. one univariate density for each dimension in the observation variable and each state). As shown in the next table, densities are recycled to simplify user-input code.
| R | User input | Density | Action |
|---|---|---|---|
| 1 | 1 | UnivariateDensity | repeat_K |
| 1 | K | UnivariateDensity | repeat_none |
| >1 | 1 | MultivariateDensity | repeat_K |
| >1 | 1 | UnivariateDensity | repeat_KxR |
| >1 | K | MultivariateDensity | repeat_none |
| >1 | K | UnivariateDensity | repeat_R |
| >1 | R | UnivariateDensity | repeat_K |
| >1 | KxR | UnivariateDensity | repeat_none |
For a univariate model (\(R = 1\)):
- User enters one univariate density: specify the same density in each hidden state variable.
- User enters \(K\) univariate density: specify one different density in each hidden state (ex. Gaussian in one state and Student in the other).
This system is flexible enough to let the user specify different densities and priors for each state. The following example demonstrates a (rather unrealistic) model where states are a priori believed to have negative, near zero, and positive location parameters.
mySpec <- hmm(
K = 3, R = 1,
observation =
Gaussian(mu = -10, sigma = 10) +
Gaussian(mu = 0, sigma = 10) +
Gaussian(mu = 10, sigma = 10),
initial = Dirichlet(alpha = ImproperUniform()),
transition = Dirichlet(alpha = ImproperUniform()),
name = "Univariate Gaussian"
)For a multivariate model (\(R > 1\)):
- User enters one univariate density: same density for each dimension of the observation variable in all states (i.e. same density and priors for the R variables in the K latent states).
- User enters one multivariate density: same density for the observation vector in all hidden states.
- User enters \(K\) univariate density: each dimension of the observation variable has the same specification within a each state.
- User enters \(K\) multivariate density: specify a multivariate density for the observation vector for each state.
- User enters \(R\) univariate density: specify one density for each element of the observation vector, which is the same across state.
- User enters \(R \times K\) univariate density: specify density for each element of the observation vector in each state. Order is \(k_1 r_1, \dots, k_1, r_R, k_2r_1, \dots, k_2r_R, \dots, k_Kr_1, \dots, k_K,r_R\).
- When \(K = R\), \(K\) prevails.
Transition model
The user may enter a Density S3 object such as a Dirichlet, or a LinkDensity S3 object (inherits from Density) such as TransitionSoftmax(). The latter allows to specify priors for the transition regression parameter vector. For example, the following example sets regularizing priors near zero on the regression coefficients (because softmax coefficients are location invariant, we use priors to better inform the model). Note that this model is hard to identify, although we achieved satisfactory results optimizing the posterior density (i.e. maximum a posterior estimates).
mySpec <- hmm(
K = 2, R = 1,
observation = Gaussian(
mu = Gaussian(0, 10),
sigma = Student(mu = 0, sigma = 10, nu = 1, bounds = list(0, NULL))
),
initial = ImproperUniform(),
transition = TransitionSoftmax(
uBeta = Gaussian(0, 5)
),
name = "TVHMM Softmax Univariate Gaussian"
)Internally, a specification stores either \(K\) multivariate densities (i.e. one multivariate density for each row in the transition matrix) or \(K \times K\) univariate densities (i.e. one univariate density for each element in the transition matrix). Densities are recycled accordingly.
| User-input | Density | Action |
|---|---|---|
| 1 | UnivariateDensity | repeat_KxK |
| 1 | MultivariateDensity | repeat_K |
| 1 | LinkDensity | nothing |
| K | UnivariateDensity | repeat_K |
| K | MultivariateDensity | repeat_none_multivariate |
| KxK | UnivariateDensity | repeat_none_univariate |
NOTE: As of the time of this writing, fixed values for transition parameters have not been implemented yet.
Initial model
Internally, a specification stores either one multivariate densities for the whole initial probability vector, or \(K\) univariate densities for each element of said vector. Densities are recycled accordingly.
| User-input | Density | Action |
|---|---|---|
| 1 | UnivariateDensity | repeat_K |
| 1 | MultivariateDensity | repeat_none_multivariate |
| 1 | LinkDensity | repeat_none_multivariate |
| K | UnivariateDensity | repeat_none_univariate |
mySpec <- hmm(
K = 2, R = 1,
observation = Gaussian(
mu = Gaussian(0, 10),
sigma = Student(mu = 0, sigma = 10, nu = 1, bounds = list(0, NULL))
),
initial = InitialSoftmax(
vBeta = Gaussian(0, 5)
),
transition = Dirichlet(alpha = ImproperUniform()),
name = "Univariate Gaussian with covariates for initial model"
)2. Validate
Bayesian Software Validation allows users to test software accuracy [CITE]. We provide a one-liner to run a validation protocol based on the prior predictive density. Iterations are run in parallel using foreach and doParallel.
Validation protocol inspired by Simulation Based Calibration (cite)
- Compile the prior predictive model (i.e. no likelihood statement in the Stan code).
- Draw \(N\) samples of the parameter vector \(\theta\) and the observation vector \({\mathbf{y}}_t\) from prior predictive density.
- Compile the posterior predictive model (i.e. Stan code includes both prior density and likelihood statement).
- For all \(n \in 1, \dots, N\):
- Feed \({\mathbf{y}}_t^{(n)}\) to the full model.
- Draw one posterior sample of the observation variable \({\mathbf{y_t}}^{(n)}_{\text{new}}\).
- Collect Hamiltonian Monte Carlo diagnostics: number of divergences, number of times max tree depth is reached, maximum leapfrogs, warm up and sample times.
- Collect posterior sampling diagnostics: posterior summary measures (mean, sd, quantiles), comparison against the true value (rank), MCMC convergence measures (Monte Carlo SE, ESS, R Hat).
- Collect posterior predictive diagnostics: observation ranks, Kolmogorov-Smirnov statistic for observed sample vs posterior predictive samples.
- Other: we expect to enrich the protocol with more relevant quantities in future iterations.
There is no such a thing as an universal, automatic way to validate models. The software computes and summarizes different diagnostic measures and provides the user with high-level tools to assess calibration (including printouts, tables, and plots). Because the user may want to single out and inspect one of the iterations, in future versions it will be able to recover (or reproduce) the full posterior sample drawn in the \(n\)-th iteration.
In the following example, we specify a HMM and validate its calibration.
mySpec <- hmm(
K = 3, R = 2,
observation =
Gaussian(mu = Gaussian(mu = -10, sigma = 1), sigma = 1) +
Gaussian(mu = Gaussian(mu = 0, sigma = 1), sigma = 1) +
Gaussian(mu = Gaussian(mu = 10, sigma = 1), sigma = 1),
initial = Dirichlet(alpha = c(0.5, 0.5, 0.5)),
transition =
Dirichlet(alpha = c(1.0, 0.2, 0.2)) +
Dirichlet(alpha = c(0.2, 1.0, 0.2)) +
Dirichlet(alpha = c(0.2, 0.2, 1.0)),
name = "Dummy Model"
)
val <- validate_calibration(mySpec, N = 2, T = 100, iter = 500, seed = 9000)
#> hash mismatch so recompiling; make sure Stan code ends with a blank line
#> hash mismatch so recompiling; make sure Stan code ends with a blank lineThe function returns a named list with two elements, chains and parameters, which are data frames storing the aforementioned summary quantities.
knitr::kable(
head(val$chains),
digits = 2, caption = "Chains diagnostics"
)| model | chain | divergences | maxTreeDepth | maxNLeapfrog | warmup | sample | r1.y25Rank | r1.yMedRank | r1.y75Rank | r1.yMeanRank | r1.ksMean | r1.ksMedian | r2.y25Rank | r2.yMedRank | r2.y75Rank | r2.yMeanRank | r2.ksMean | r2.ksMedian | seed | n |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dummy Model | 1 | 33 | 0 | 25 | 0.76 | 0.52 | 0.12 | 0.61 | 0.82 | 0.35 | 0.29 | 0.30 | 0.90 | 0.80 | 0.65 | 0.67 | 0.22 | 0.21 | 9000 | 1 |
| Dummy Model | 2 | 0 | 0 | 15 | 0.81 | 0.55 | 0.02 | 0.14 | 0.74 | 0.17 | 0.34 | 0.35 | 0.83 | 0.80 | 0.57 | 0.62 | 0.23 | 0.22 | 9000 | 1 |
| Dummy Model | 3 | 0 | 0 | 15 | 0.84 | 0.52 | 0.01 | 0.17 | 0.41 | 0.11 | 0.37 | 0.37 | 0.80 | 0.76 | 0.70 | 0.65 | 0.22 | 0.22 | 9000 | 1 |
| Dummy Model | 4 | 0 | 0 | 31 | 0.98 | 0.54 | 0.02 | 0.03 | 0.45 | 0.00 | 0.34 | 0.34 | 0.79 | 0.78 | 0.59 | 0.60 | 0.23 | 0.22 | 9000 | 1 |
| Dummy Model | 1 | 0 | 0 | 31 | 1.23 | 0.54 | 0.05 | 0.02 | 0.61 | 0.03 | 0.50 | 0.53 | 0.00 | 0.13 | 0.68 | 0.48 | 0.23 | 0.22 | 9000 | 2 |
| Dummy Model | 2 | 0 | 0 | 15 | 1.11 | 0.62 | 0.02 | 0.00 | 0.24 | 0.01 | 0.50 | 0.52 | 0.00 | 0.09 | 0.82 | 0.53 | 0.23 | 0.22 | 9000 | 2 |
knitr::kable(
head(val$parameters),
digits = 2, caption = "Parameters diagnostics"
)| model | chain | parameter | mean | se_mean | sd | X2.5. | X25. | X50. | X75. | X97.5. | n_eff | Rhat | true | rank | n |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dummy Model | 1 | mu11 | -10.10 | 0.10 | 1.14 | -12.15 | -10.96 | -10.04 | -9.26 | -8.13 | 125.00 | 1.01 | -11.47 | 0.10 | 1 |
| Dummy Model | 1 | mu12 | -10.64 | 0.01 | 0.09 | -10.84 | -10.68 | -10.64 | -10.58 | -10.47 | 125.00 | 0.99 | -9.91 | 1.00 | 1 |
| Dummy Model | 1 | mu21 | -0.03 | 0.10 | 1.01 | -1.87 | -0.77 | -0.12 | 0.70 | 2.00 | 92.91 | 0.99 | -0.46 | 0.34 | 1 |
| Dummy Model | 1 | mu22 | 9.74 | 0.01 | 0.13 | 9.47 | 9.67 | 9.75 | 9.81 | 9.96 | 125.00 | 0.99 | 0.75 | 0.00 | 1 |
| Dummy Model | 1 | mu31 | 9.91 | 0.12 | 0.95 | 8.23 | 9.28 | 9.72 | 10.58 | 11.98 | 62.18 | 1.00 | 10.48 | 0.74 | 1 |
| Dummy Model | 1 | mu32 | 0.57 | 0.01 | 0.15 | 0.28 | 0.47 | 0.58 | 0.68 | 0.85 | 125.00 | 1.00 | 9.60 | 1.00 | 1 |
plot(
val$parameters$Rhat,
type = "h", ylim = c(0.9, 1.15),
ylab = expression(hat(R)),
xlab = "Validation run (n)"
)
abline(h = 1.1)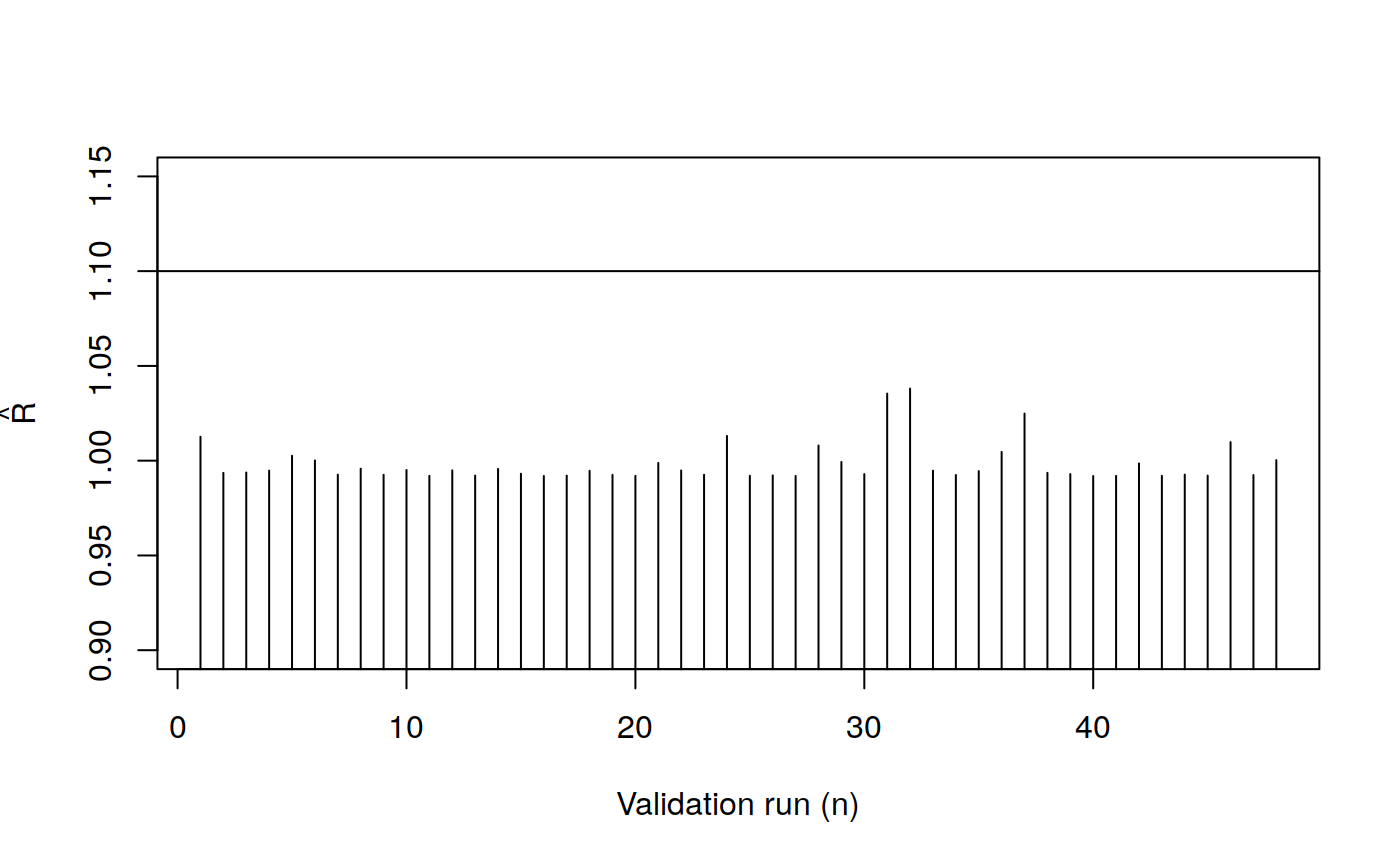
3. Simulate & Fit
Once specified, the user may:
- Call
sim()to simulate \(N\) datasets containing the observation variables \({\mathbf{y}}\) and the hidden paths \(z\). - Call
compile()to compile the underlying Stan model. The returned object may be later used for either sampling (MCMC) or optimization (MAP).- Call
sampling()to draw posterior samples from a compiled model effectively allowing for full Bayesian inference via MCMC. - Call
optimizing()to maximize the joint posterior density and obtain a point estimate.
- Call
- Call
fit()to compile and sample in one step.
After fitting, the underlying Stan code may be inspected by calling browse_model, which opens the Stan file in the editor. None that this function is only for reading purposes: changes in the Stan code will not affect the specification object.
3.1. Fully Bayesian Inferencia via MCMC
Because sampling() and fit() return a stanfit S4 object, the posterior samples are compatible out of box with the rstan ecosystem [TRY AND LIST SOME SUCH AS bayesplot].
3.2. Maximum a Posteriori estimates via posterior optimization
optimizing is able to run several initialization of the optimization algorithm in parallel. The user may decide to keep = "all" runs, or just keep = "best". If the former, extract_grid returns a summary of the different runs and extract_best retrieves the one with highest posterior log likelihood. If no run achieves convergence, a warning message is prompt to the console.
mySpec <- hmm(
K = 3, R = 1,
observation =
Gaussian(mu = Gaussian(mu = -10, sigma = 1), sigma = 1) +
Gaussian(mu = Gaussian(mu = 0, sigma = 1), sigma = 1) +
Gaussian(mu = Gaussian(mu = 10, sigma = 1), sigma = 1),
initial = Dirichlet(alpha = c(0.5, 0.5, 0.5)),
transition = Dirichlet(alpha = c(0.5, 0.5, 0.5)),
name = "Univariate Gaussian"
)
set.seed(9000)
y <- as.matrix(
c(rnorm(100, -10, 1), rnorm(100, 0, 1), rnorm(100, 10, 1))
)
y <- sample(y)
myModel <- compile(mySpec)
myAll <- optimizing(mySpec, myModel, y = y, nRun = 10, keep = "all", nCores = 4)
myBest <- extract_best(myAll)print(
round(extract_grid(myAll, pars = "mu"), 2)
)
#> n seed logPosterior returnCode mu11 mu21 mu31
#> [1,] 8 1111034741 -820.72 0 0 0 0 -10.06 9.98 0.04
#> [2,] 2 768003877 -820.72 0 0 0 0 -10.06 9.98 0.04
#> [3,] 6 820771557 -820.72 0 0 0 0 -10.06 9.98 0.04
#> [4,] 10 2115105852 -820.72 0 0 0 0 -10.06 9.98 0.04
#> [5,] 5 1539977502 -1018.84 0 0 0 0 -0.16 9.98 -9.87
#> [6,] 9 355282361 -1018.84 0 0 0 0 -0.16 9.98 -9.86
#> [7,] 7 2066428059 -1020.17 0 0 0 0 9.88 -9.96 0.04
#> [8,] 3 1898572606 -1020.17 0 0 0 0 9.88 -9.96 0.04
#> [9,] 1 1144725567 -1119.23 0 0 0 0 9.88 -0.06 -9.87
#> [10,] 4 433311989 -1119.23 0 0 0 0 9.88 -0.06 -9.87NOTE: Passing both the specification and the compiled model is redundant. In future versions, user will not need to pass the specification.
3.3 Common high-level routines
Most of these extractors rely on the lower level extract_quantity function. By default, it returns a named list where each element is an array representing a sampled quantity. The dimension of the array varies according to the number of iterations, the length of the series, and the structure of the quantity itself (for example, whether they are univariate or multivariate). Additionally, the function accepts wildcards * for easy extraction of a family of quantity.
This function has two convenience arguments:
-
reducetakes a function that is run with-in chain, effectively reducing the \(N\) draws per chain into one or more quantity. -
combinetakes a function that is run on the reduced quantity for each parameter, effectively combining the summary quantity from each parameter into a single data structure.
Besides extract_quantity, which takes an argument pars, the software comes with extractors for the most typical quantities: extract_alpha (filtered probability), extract_gamma (smoothed probability), extract_zstar (MPM, also known a Viterbi), extract_ypred and extract_zpred for predictive quantities, and extract_obs_parameters and extract_parameters for observation model and all model parameters respectively.
NOTE: Naming convention may be changed before submitting to CRAN. Consider for example extract_alpha versus extract_filtered_probability, or extract_zstar versus extract_viterbi.
Compare in these examples different configurations of reduce and combine :
mySpec <- hmm(
K = 3, R = 1,
observation =
Gaussian(mu = Gaussian(mu = -10, sigma = 1), sigma = 1) +
Gaussian(mu = Gaussian(mu = 0, sigma = 1), sigma = 1) +
Gaussian(mu = Gaussian(mu = 10, sigma = 1), sigma = 1),
initial = Dirichlet(alpha = c(0.5, 0.5, 0.5)),
transition = Dirichlet(alpha = c(0.5, 0.5, 0.5)),
name = "Univariate Gaussian"
)
set.seed(9000)
y <- as.matrix(
c(rnorm(100, -10, 1), rnorm(100, 0, 1), rnorm(100, 10, 1))
)
y <- sample(y)
myFit <- fit(mySpec, y = y, iter = 500)
# 3 parameters, 1,000 iterations, 4 chains
print(
str(extract_obs_parameters(myFit))
)
#> List of 3
#> $ mu11: num [1:250, 1:4] 9.73 10.1 10.05 9.8 9.95 ...
#> $ mu21: num [1:250, 1:4] -0.1933 -0.189 -0.1924 0.0366 -0.0616 ...
#> $ mu31: num [1:250, 1:4] -9.93 -9.84 -9.82 -9.88 -9.77 ...
#> NULL
# reduce within-chain draws to one quantity (median)
print(
extract_obs_parameters(myFit, reduce = median)
)
#> $mu11
#> [1] 9.8797135 -0.1670912 -0.1545372 9.8801167
#>
#> $mu21
#> [1] -0.04011546 9.98843546 9.98835770 -9.96346881
#>
#> $mu31
#> [1] -9.8604877 -9.8669435 -9.8609835 0.0377377
# reduce within-chain draws to two quantities (quantiles)
print(
extract_obs_parameters(
myFit, reduce = posterior_intervals(c(0.1, 0.9))
)
)
#> $mu11
#> [,1] [,2] [,3] [,4]
#> [1,] 9.749743 -0.28719452 -0.25653428 9.742132
#> [2,] 10.008315 -0.03632407 -0.04306187 10.014145
#>
#> $mu21
#> [,1] [,2] [,3] [,4]
#> [1,] -0.19350995 9.856644 9.852452 -10.100755
#> [2,] 0.07658331 10.104012 10.112068 -9.824095
#>
#> $mu31
#> [,1] [,2] [,3] [,4]
#> [1,] -10.002818 -9.992977 -9.990071 -0.09496092
#> [2,] -9.722103 -9.741113 -9.745545 0.17199537
# combine quantities into a matrix
print(
extract_obs_parameters(
myFit,
reduce = median,
combine = rbind
)
)
#> [,1] [,2] [,3] [,4]
#> mu11 9.87971351 -0.1670912 -0.1545372 9.8801167
#> mu21 -0.04011546 9.9884355 9.9883577 -9.9634688
#> mu31 -9.86048774 -9.8669435 -9.8609835 0.0377377Since classification is a frequent task in the data analysis workflow, we provide a family of classify_* functions that extract the estimated quantities and return the most likely state: classify_alpha and classify_gamma return the state with highest posterior probability at each time step, while classify_zstar returns the posterior mode of the jointly most likely state.
print(
str(classify_alpha(myFit))
)
#> int [1:300, 1:4] 3 2 2 1 1 2 2 1 1 2 ...
#> NULL
print(
str(classify_gamma(myFit))
)
#> int [1:300, 1:4] 3 2 2 1 1 2 2 1 1 2 ...
#> NULL
print(
str(classify_zstar(myFit))
)
#> num [1:4, 1:300] 3 3 3 2 2 1 1 3 2 1 ...
#> NULL4. Diagnose
This step is not implemented as of the date of this writing. We plan to explore how to diagnose a HMM outside posterior predictive checks (think, for example, pseudo residuals).
5. Visualize
We expect that visualization will play a very important role in our software once it arrives to a mature stage. At this point, we support a small amount of graphical routines:
-
plot_seriesfocuses on the observation variables. -
plot_state_probabilityfocuses on the estimated assignment probability. -
plot_ppredictivefocuses on posterior predictive checks.
Besides functions for fully-customizable individual plots such as plot_ppredictive_density(y, yPred) (note that the function takes as arguments the actual values and not a fitted object), the software includes a convenient way to automatically include one or more plots from the same family on a single layout. Each plot may toggle one or more “feature” such as colored lines, points, or backgrounds. Additionally, plots adjust automatically for multivariate observations, and variable names are automatically shown if the data matrix used to fitted the model has named columns. Colors are themeable via global options.
The following features are currently available:
When plotting the observation variables: *
stateShadeto shade the background based on the state probability. *yColoredLineto include a line colored according to the state probability. *yColoredDotsto include dots colored according to the state probability.When plotting the state probabilities: *
stateShadeto shade the background based on the state probability. *probabilityColoredLineto include a line colored according to the state probability. *probabilityColoredDotsto include dots colored according to the state probability. *bottomColoredMarksandtopColoredMarksto include colored tick marks on the bottom or top axis. *probabilityFanto show the posterior interval of assignment probabilities.
Again, the objects returned by fit and sampling are compatible with the rstan ecosystem, including visualization routines in the bayesplot package.
mySpec <- hmm(
K = 3, R = 2,
observation =
Gaussian(mu = Gaussian(mu = -10, sigma = 1), sigma = 1) +
Gaussian(mu = Gaussian(mu = 0, sigma = 1), sigma = 1) +
Gaussian(mu = Gaussian(mu = 10, sigma = 1), sigma = 1),
initial = Dirichlet(alpha = c(0.5, 0.5, 0.5)),
transition =
Dirichlet(alpha = c(1.0, 0.2, 0.2)) +
Dirichlet(alpha = c(0.2, 1.0, 0.2)) +
Dirichlet(alpha = c(0.2, 0.2, 1.0)),
name = "Univariate Gaussian Dummy Model"
)
mySim <- sim(mySpec, T = 300, seed = 9000, iter = 500, chains = 1)
ySim <- extract_ypred(mySim)[1, , ]
colnames(ySim) <- c("Weight", "Height")
myFit <- fit(mySpec, y = ySim, seed = 9000, iter = 500, chains = 1)plot_series(myFit, legend.cex = 0.8)
plot_series(myFit, xlab = "Time steps", features = c("yColoredLine"))
plot_series(
myFit, stateProbability = "smoothed",
features = c("stateShade", "bottomColoredMarks")
)
plot_state_probability(myFit, main = "Title", xlab = "Time")
plot_state_probability(myFit, features = c("stateShade"), main = "Title", xlab = "Time")
# plot_state_probability(myFit, features = c("bottomColoredMarks"), main = "Title", xlab = "Time")
plot_state_probability(myFit, features = c("probabilityColoredDots"), main = "Title", xlab = "Time")
plot_state_probability(myFit, features = c("probabilityColoredLine"), main = "Title", xlab = "Time")
# plot_state_probability(myFit, stateProbability = "filtered", features = c("bottomColoredMarks", "probabilityFan"), stateProbabilityInterval = c(0.05, 0.95), main = "Title", xlab = "Time")
plot_ppredictive(myFit, type = c("density", "cumulative", "summary"), fun = median)
plot_ppredictive(myFit, type = c("density", "boxplot"), fun = median, subset = 1:10)
plot_ppredictive(
myFit,
type = c("density", "boxplot", "scatter"),
fun = median, fun1 = mean, fun2 = median,
subset = 1:40
)
plot_ppredictive(myFit, type = c("density", "cumulative", "hist"), fun = median)
plot_ppredictive(myFit, type = c("density", "cumulative", "ks"))Acknowledgements
We acknowledge Google Summer of Code 2018 for funding.
Appendix
We present 19 different possible configurations.
# OBSERVATION MODEL -------------------------------------------------------
K = 3
R = 2
# Case 1. A different multivariate density for each state
# Input: K multivariate densities
# Behaviour: Nothing
exCase1 <- hmm(
K = K, R = R,
observation =
MVGaussianCholeskyCor(
mu = Gaussian(mu = 0, sigma = 1),
L = LKJCor(eta = 2)
) +
MVGaussianCholeskyCor(
mu = Gaussian(mu = 0, sigma = 10),
L = LKJCor(eta = 3)
) +
MVGaussianCholeskyCor(
mu = Gaussian(mu = 0, sigma = 100),
L = LKJCor(eta = 4)
),
initial = ImproperUniform(),
transition = ImproperUniform(),
name = "A different multivariate density for each each state"
)
# Case 2. Same multivariate density for every state
# Input: One multivariate density
# Behaviour: Repeat input K times
exCase2 <- hmm(
K = K, R = R,
observation =
MVGaussianCholeskyCor(
mu = Gaussian(mu = 0, sigma = 100),
L = LKJCor(eta = 2)
),
initial = ImproperUniform(),
transition = ImproperUniform(),
name = "Same multivariate density for every state"
)
# Case 3. Same univariate density for every state and every output variable
# Input: One univariate density
# Behaviour: Repeat input K %nested% R times
exCase3 <- hmm(
K = K, R = R,
observation =
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
),
initial = ImproperUniform(),
transition = ImproperUniform(),
name = "Same univariate density for every state and every output variable"
)
# Case 4. Same R univariate densities for every state
# Input: R univariate densities
# Behaviour: Repeat input K times
exCase4 <- hmm(
K = K, R = R,
observation =
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
) +
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
),
initial = ImproperUniform(),
transition = ImproperUniform(),
name = "Same R univariate densities for every state"
)
# Case 5. Same univariate density for every output variable
# Input: K univariate densities
# Behaviour: Repeat input R times
exCase5 <- hmm(
K = K, R = R,
observation =
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
) +
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
) +
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
),
initial = ImproperUniform(),
transition = ImproperUniform(),
name = "Same univariate density for every output variable"
)
# Case 6. Different univariate densities for every pair of state and output variable
# Input: K %nested% R univariate densities
# Behaviour: None
exCase6 <- hmm(
K = K, R = R,
observation =
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
) +
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
) +
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
) +
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
) +
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
) +
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
),
initial = ImproperUniform(),
transition = ImproperUniform(),
name = "Different univariate densities for every pair of state and output variable"
)
# UNIVARIATE Observation model --------------------------------------------
K = 3
R = 1
# Case 7. A different univariate density for each each state
# Input: K univariate densities
# Behaviour: Nothing
exCase7 <- hmm(
K = K, R = R,
observation =
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
) +
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
) +
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
),
initial = ImproperUniform(),
transition = ImproperUniform(),
name = "A different univariate density for each each state"
)
# Case 8. Same univariate density for every state
# Input: One univariate density
# Behaviour: Repeat input K times
exCase8 <- hmm(
K = K, R = R,
observation =
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
),
initial = ImproperUniform(),
transition = ImproperUniform(),
name = "Same multivariate density for every state"
)
# INITIAL MODEL -----------------------------------------------------------
K = 3
R = 2
# Case 9. Same univariate density for every initial state
# Input: One univariate density
# Behaviour: Repeat input K times
exCase9 <- hmm(
K = K, R = R,
observation =
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
),
initial =
Beta(
alpha = Gaussian(0, 1),
beta = Gaussian(1, 10)
),
transition = ImproperUniform(),
name = "Same univariate density for every initial state"
)
# Case 10. One multivariate density for the whole initial vector
# Input: One multivariate density
# Behaviour: Nothing
exCase10 <- hmm(
K = K, R = R,
observation =
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
),
initial =
Dirichlet(
alpha = ImproperUniform()
),
transition = ImproperUniform(),
name = "One multivariate density for the whole initial vector"
)
# Case 11. A different univariate density for each initial state
# Input: K univariate densities
# Behaviour: Nothing
exCase11 <- hmm(
K = K, R = R,
observation =
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
),
initial =
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
) +
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
) +
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
),
transition = ImproperUniform(),
name = "A different univariate density for each initial state"
)
# Case 12. A link
# Input: One link density
# Behaviour: Nothing
exCase12 <- hmm(
K = K, R = R,
observation =
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
),
initial =
InitialSoftmax(
vBeta = ImproperUniform()
),
transition = ImproperUniform(),
name = "TV Initial distribution"
)
# TRANSITION MODEL --------------------------------------------------------
K = 3
R = 2
# Case 13. Same univariate density for every transition
# Input: One univariate density
# Behaviour: Repeat input KxK times
exCase13 <- hmm(
K = K, R = R,
observation =
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
),
initial = Dirichlet(alpha = c(0.5, 0.5, 0.5)),
transition =
Gaussian(mu = 0, sigma = 1),
name = "Same univariate density for every transition"
)
# Case 14. Same multivariate density for every transition row
# Input: One multivariate density
# Behaviour: Repeat input K times
exCase14 <- hmm(
K = K, R = R,
observation =
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
),
initial = Dirichlet(alpha = c(0.5, 0.5, 0.5)),
transition =
Dirichlet(
alpha = c(0.5, 0.5, 0.7)
),
name = "Same multivariate density for every transition row"
)
# Case 15. A different univariate density for each element of the transition row
# Input: K univariate densities
# Behaviour: Repeat input K times
exCase15 <- hmm(
K = K, R = R,
observation =
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
),
initial = Dirichlet(alpha = c(0.5, 0.5, 0.5)),
transition =
Beta(alpha = 0.1, beta = 0.1) +
Beta(alpha = 0.5, beta = 0.5) +
Beta(alpha = 0.9, beta = 0.9),
name = "A different univariate density for each element of the transition row"
)
# Case 16. A different multivariate density for each transition row
# Input: K multivariate densities
# Behaviour: nothing
exCase16 <- hmm(
K = K, R = R,
observation =
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
),
initial = Dirichlet(alpha = c(0.5, 0.5, 0.5)),
transition =
Dirichlet(alpha = c(0.1, 0.1, 0.1)) +
Dirichlet(alpha = c(0.5, 0.5, 0.5)) +
Dirichlet(alpha = c(0.9, 0.9, 0.9)),
name = "A different multivariate density for each transition row"
)
# Case 17. Different univariate densities for each element of the transition matrix
# Input: KxK univariate densities
# Behaviour: None
exCase17 <- hmm(
K = K, R = R,
observation =
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
),
initial = Dirichlet(alpha = c(0.5, 0.5, 0.5)),
transition =
Beta(alpha = 0.1, beta = 0.1) +
Beta(alpha = 0.2, beta = 0.2) +
Beta(alpha = 0.3, beta = 0.3) +
Beta(alpha = 0.4, beta = 0.4) +
Beta(alpha = 0.5, beta = 0.5) +
Beta(alpha = 0.6, beta = 0.6) +
Beta(alpha = 0.7, beta = 0.7) +
Beta(alpha = 0.8, beta = 0.8) +
Beta(alpha = 0.9, beta = 0.9),
name = "Different univariate densities for each element of the transition matrix"
)
# Case 18. A link
# Input: One link density
# Behaviour: Nothing
exCase18 <- hmm(
K = K, R = R,
observation =
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
),
initial = Dirichlet(alpha = c(0.5, 0.5, 0.5)),
transition =
TransitionSoftmax(
uBeta = Gaussian(mu = 0, sigma = 1)
),
name = "Different univariate densities for each element of the transition matrix"
)
# A FULLY COMPLEX MODEL ---------------------------------------------------
# Case 19. A link in both the transition and initial distribution.
# Input: A link in both the transition and initial distribution.
# Behaviour: Nothing
exCase19 <- hmm(
K = K, R = R,
observation =
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
) +
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
) +
Gaussian(
mu = Gaussian(0, 10),
sigma = Gaussian(0, 10, bounds = list(0, NULL))
),
initial =
InitialSoftmax(
vBeta = Gaussian(mu = 0, sigma = 1)
),
transition =
TransitionSoftmax(
uBeta = Gaussian(mu = 0, sigma = 1)
),
name = "Fully Complex Model"
)